This blog will discuss the various DNA Processes required for DNA(replication), RNA(transcription), and Protein(translation) synthesis, more focused on replication.
As we know, DNA is the evolved form of RNA and follows all the properties required for the material to be recognized as genetic material. The properties followed by the DNA are in different aspects like it can self-replicate, it is highly stable, it allows mutations, and it is also expressed in mendelian characters. We will discuss in more detail mendelian characters in upcoming blogs. Like how mendelian genetics comes forward, how it exactly works, and also in detail about its discovery and laws.
Coming to the main discussion today, we will discuss all the properties of genetic material in detail. As we said, DNA can self-replicate, but how? It can adapt mutations, but how? And it also can form a stable structure and store genetic information, but how? DNA can form DNA, RNA, and Protein. These functional abilities make DNA a superman in the human body who can control all the synthesis of essential biomolecules like proteins and amino acids.

The DNA process involves the synthesis of DNA from DNA is named replication in that DNA form an exact copy of itself using various other helping hand reagents. The formation of DNA to RNA is called transcription, and the synthesis of Protein from RNA is termed translation. Similarly, if DNA is formed from RNA is known as reverse transcription.
DNA Replication
Replication is the essential property required for the material to be genetic. According to this, the material can be self-replicated. Watson and Crick proposed this scheme for the replication of DNA. It suggests that the two strands of DNA would separate and act as a template for synthesizing the new complementary strand.
We have already discussed what is exactly the template strand, coding strand, and complementary strand. Replication in easy language is known as duplication of formation of the replica. This replication process is worked in three phases, namely initiation, elongation, and termination. Various chemical agents are used in all this separation process, uncoiling, cutting and recoiling, and again separating new strands.
Now the fundamental question is why we are discussing replication and what is the need for DNA replication? So, we know cells are continuously dividing in our body, and as we know in eukaryotes, DNA is stored in the cell’s nucleus. When a cell divides and forms its daughter cells, DNA replicates and forms its two replicas and is transported into daughter cells. This cycle of replication continues till the cell division continues.
Because every dividing cell has its fate for which it is determined, and that determination is supported by the genetic material present in that cell. Let’s discuss the replication phases.
Uncoiling of DNA (Initiation)
DNA is a double-stranded helix molecule made up of nucleotides. These double strands are attached tightly through the Hydrogen bond. During uncoiling or unwinding, these H-bonds partially break, and the Separation of Double Helix occurs. During cell, division cells undergo a cell cycle mitosis during the synthesis (S) phase of cell cycle replication.
In prokaryotes and eukaryotes, this replication starts on a particular site only that is the replication site. OriC– site that is the origin of replication in many organism-like prokaryotes and eukaryotes.
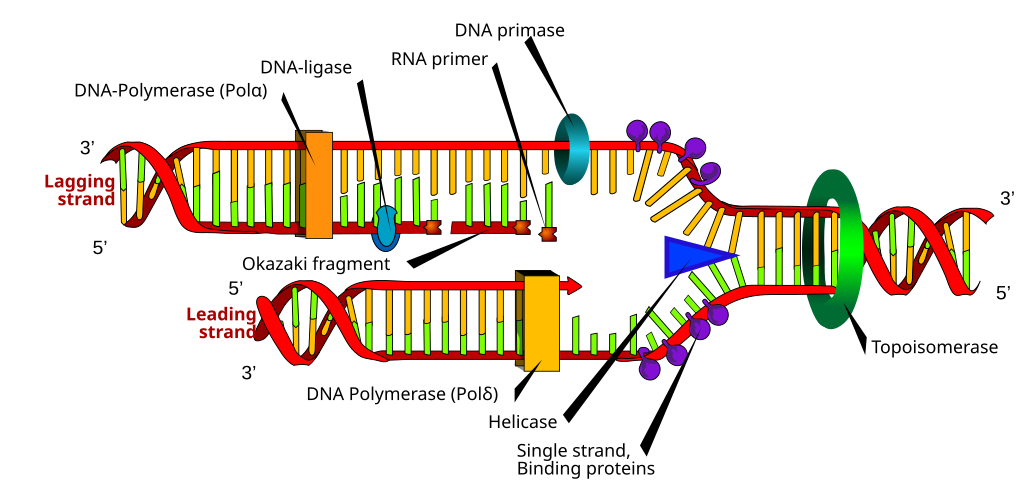
The Oric site is an adenine- thymine (A=T) rich region where the helicase enzyme is bound. The binding of the helicase enzyme leads to the breaking of H-bonds and forms a bubble or replication fork. Replication is a bidirectional process; hence it works in both directions and terminates at one point. As the helicase enzyme goes forward, the separation of DNA double-strand starts. The SSB (single-stranded binding) proteins bind on separated strands to avoid recoiling and stabilize the separated single strands. DNA topoisomerases bind on the double-strand and resolve the supercoiling that occurred due to the unwinding of DNA.
Complementary Strand Synthesis(Elongation)
Immediate after the separation of double-helix DNA polymerase III enzyme binds on the 3’-OH free end of DNA and starts adding free complementary nucleotides. The nucleotides added are about the template strand. DNA polymerase enzyme has specific requirements like a free 3’-OH end and a template strand.
Two strands are formed during replication, namely the leading strand and the lagging strand.RNA primers synthesized by the primase enzyme bind on the lagging strand and provide a free 3’-OH end extended by the polymerase III enzyme. Leading strand synthesis is continuous and fast in 5’-3′ direction, but on the other hand, lagging strand synthesis is discontinuous and slow.
On the lagging strand, the extended RNA primers are termed as Okazaki fragments are formed and are joined by DNA ligase (a molecular glue). Okazaki fragments are short oligomeric DNA sequences.
The newly synthesized strand is complementary to the template strand, and as a new strand is synthesized, it forms helical binding and regains the double-helical structure.
Separation of Daughter DNA(Termination)
After finishing the replication of complete parent DNA, the newly formed daughter DNA gets separated and transported into two daughter cells. Before transport, RNA primers are removed using a special RNA cutter that is a helicase enzyme called RNase-H and the Okazaki fragments get ligated with ligase.
During the whole process, if the wrong nucleotides get added during the whole process, the DNA polymerase enzyme has a remarkable ability of correction called proofreading activity. Because of proofreading activity, DNA polymerase is more accurately working than RNA polymerase. The synthesis direction is 5′ to 3′, and proofreading can be in 3’-5′ direction, which is known as exogenous activity. The mode of this type of replication is proved to be semiconservative.
The semiconservative replication mode means the parental DNA is distributed in fifty-fifty concentrations in both daughter cells. The detailed experiment was performed on the semiconservative model of DNA. We will discuss that experiment in detail in the upcoming article.